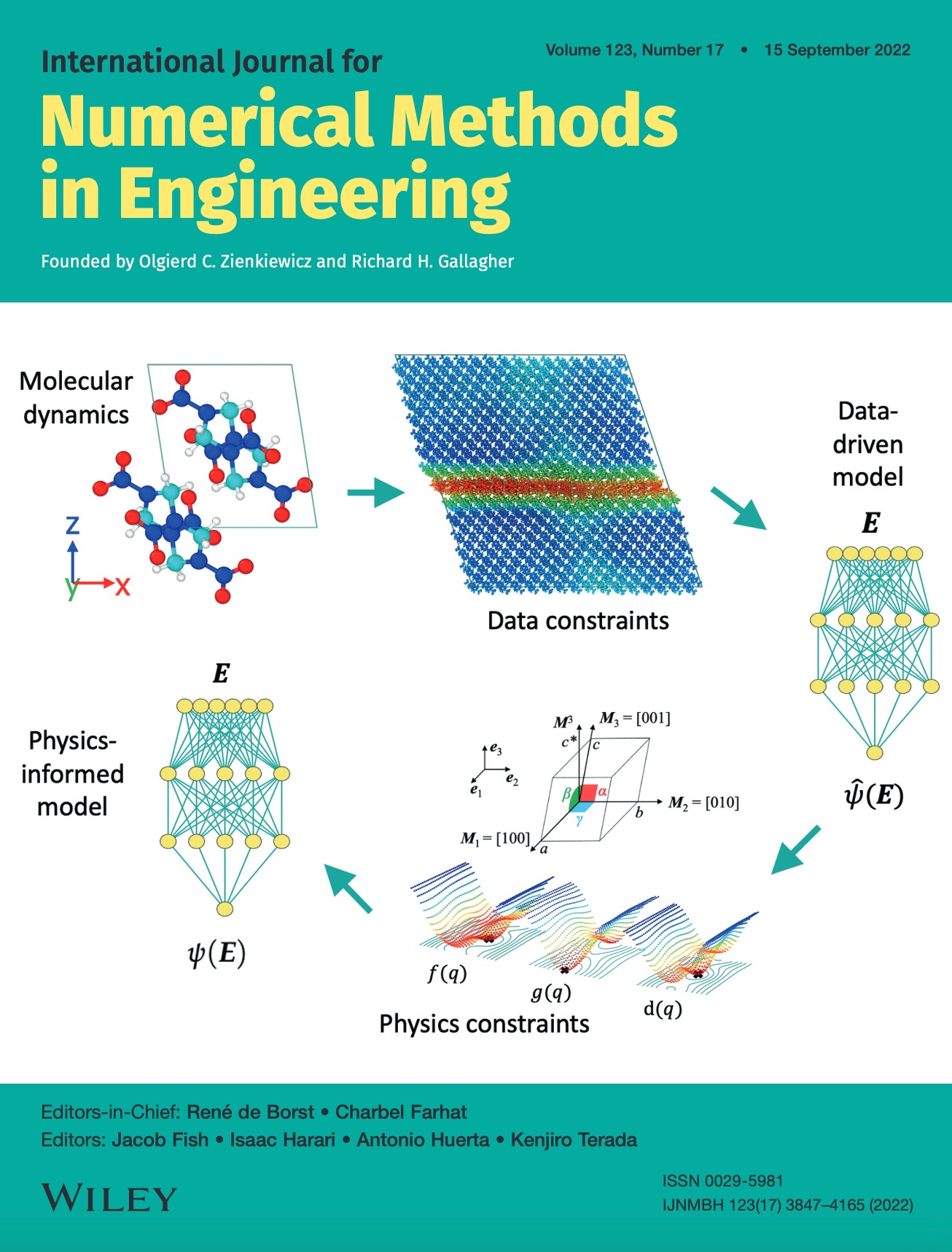
Molecular Dynamics Inferred Transfer Learning Models for Finite-Strain Hyperelasticity of Monoclinic Crystals: Sobolev Training and Validations Against Physical Constraints
Vlassis, Zhao, Ma, Sewell, Sun, IJNME, 2022.
We present a machine learning framework to train and validate neural networks to predict the anisotropic elastic response of a monoclinic organic molecular crystal known as β-HMX in the geometrical nonlinear regime. A filtered molecular dynamic (MD) simulations database is used to train neural networks with a Sobolev norm that uses the stress measure and a reference configuration to deduce the elastic stored free energy functional. To improve the accuracy of the elasticity tangent predictions originating from the learned stored free energy, a transfer learning technique is used to introduce additional tangential constraints from the data while necessary conditions (e.g., strong ellipticity, crystallographic symmetry) for the correctness of the model are either introduced as additional physical constraints or incorporated in the validation tests. Assessment of the neural networks is based on (1) the accuracy with which they reproduce the bottom-line constitutive responses predicted by MD, (2) the robustness of the models measured by detailed examination of their stability and uniqueness, and (3) the admissibility of the predicted responses with respect to mechanics principles in the finite-deformation regime. We compare the training efficiency of the neural networks under different Sobolev constraints and assess the accuracy and robustness of the models against MD benchmarks for β-HMX.
Read more →
Read more →